|
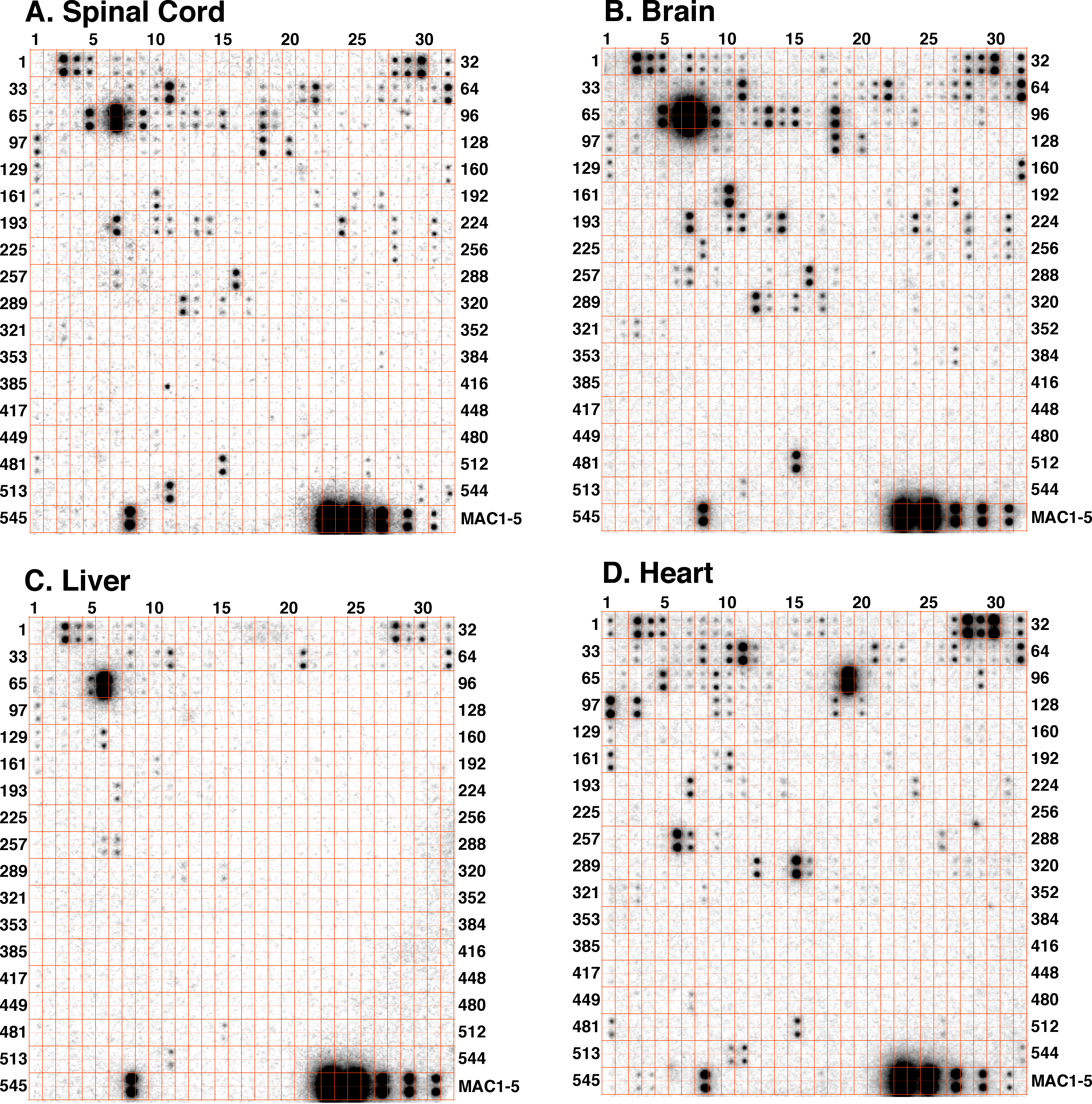
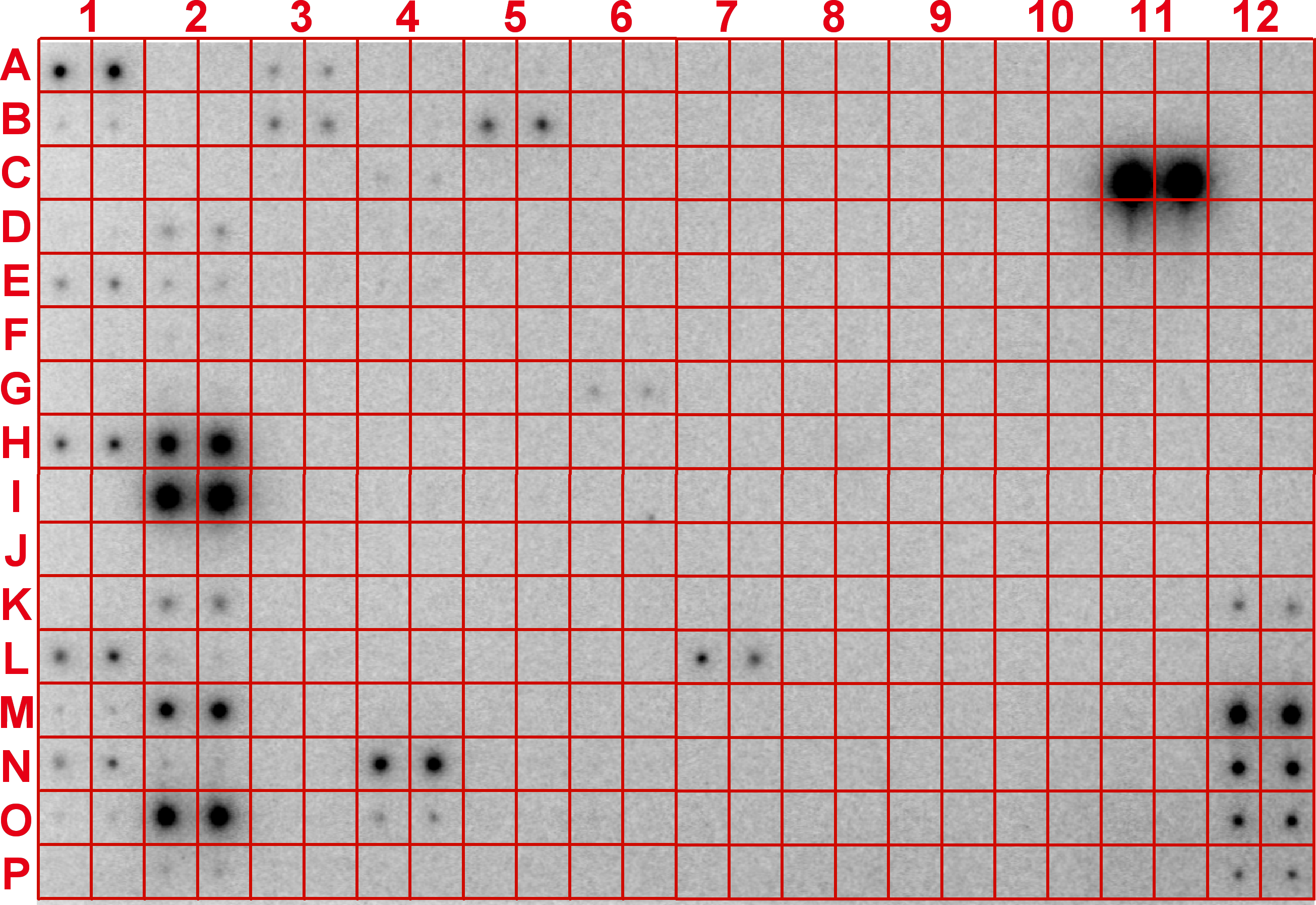
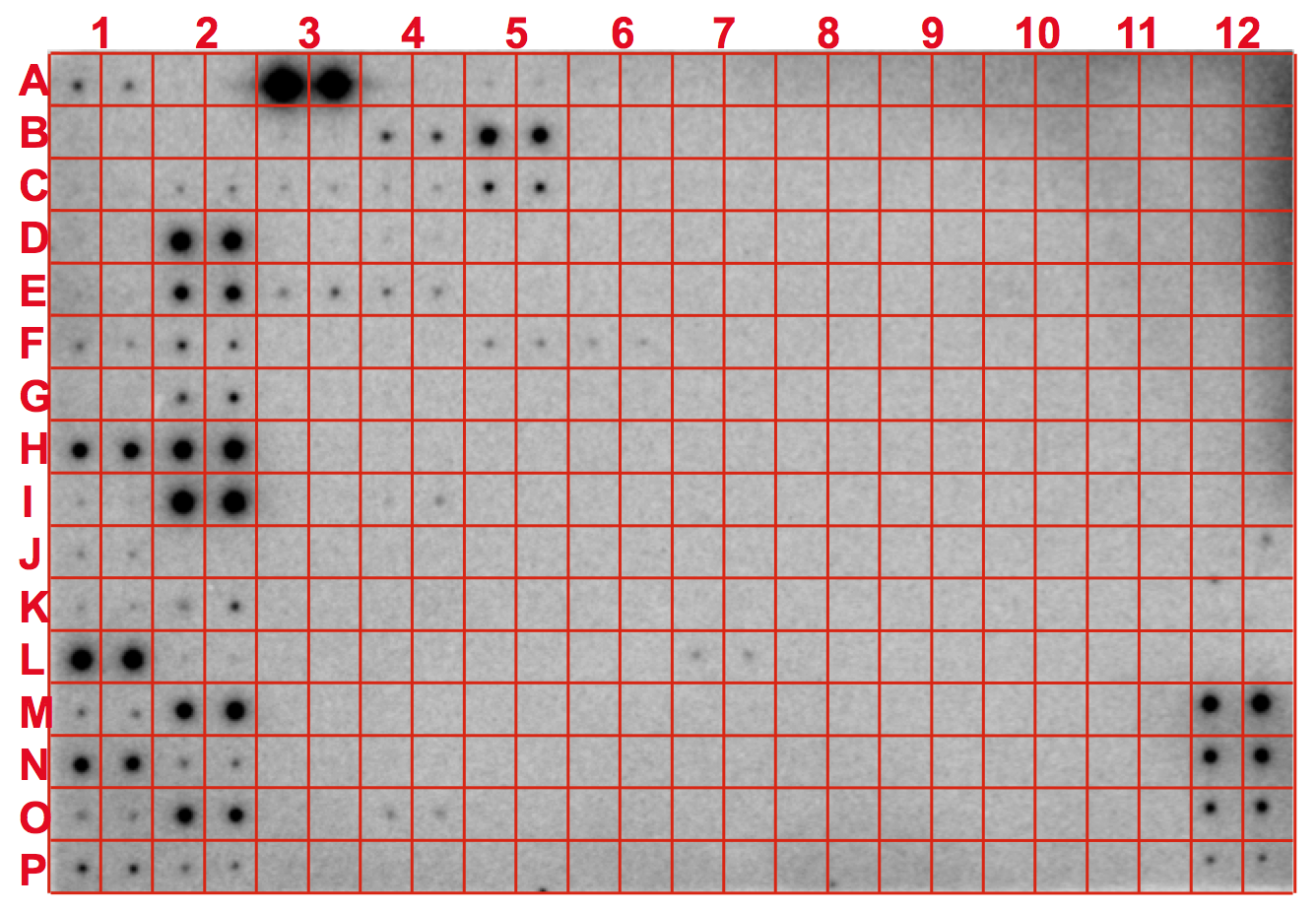
Introduction: MicroRNAs (miRNAs) are a novel class of small non-coding RNAs that regulate gene expression at the post-transcriptional level and play a critical role in many important biological processes in animals including humans. Most miRNAs are conserved between humans and mice, which makes it possible to analyze their expression patterns with a set of selected array probes. The maps of expression of selected miRNAs from different species, organs/tissues, or cell lines can be tentatively termed "microRNA atlas". Similarly in plants, most miRNAs so far identified are very conserved between species although the expression patterns of these conserved miRNAs are different between plant species, organs/tissues, or cell lines. In mammals, miRNA expression atlas as a powerful resource has recently been drawn based on small RNA library sequencing (Cell, Vol 129, 1410-1414, 29 June 2007). With different samples to be studied, the miRNA atlas will be continuiously enlarged. To simplify the mapping process and to reduce the cost of drafting a miRNA atlas for specific species, organs, tissues, or cell types, we employed our recently established simple array platform (RNA, Vol 13, 1803-1822, 3 August 2007) to draft a framework of miRNA atlas for humans/mice and plants as demonstrated below. The miRNA atlas is based on the endogenous miRNAs with no amplification, truely reflecting the in vivo levels and changes of miRNAs. We believe that this simple miRNA atlas will be a useful guider to the study of miRNAs and their functions in plants and animals. With a single miRNA atlas for your specific, normal organs/tissues/cells, you may start to study roles of miRNAs without further array for more samples of the same materials from different resources by probing those miRNAs highlighted on the miRNA atlas with traditional Northern blot analysis. For example, totally 38 miRNAs were highlighted in liver organ, miR122a is highly abundant, no miR124a that is highly expressed in central nerve system was detectable in liver. Therefore, you could synthesize 38 probes specifically for liver expressed miRNAs without need of addressing other miRNAs, such as miR124a, to study the functions of the 38 miRNAs, including the highly expressed miR122a, in liver, simply using the traditional Northern blot analysis. (To draw a miRNA atlas for your species, please contact us by email: gtang1@mtu.edu) A. Mouse miRNA atlas:
|