Simple Neural Network Classifiers
Artificial Neural Networks have been used by researchers since at least the 1950s, and rose in prominence when computing resources became widely available. There have been at least 3-4 eras of ANN, with each success and exuberance followed by a quiescence and failure or replacement by other tools. There has always been an intetrplay in this area between computational neuroscientists, computational vision, machine learning, psychology, computer science, and statisticians, with many advances coming by modeling human processes and then applying these algorithms to real-life problems.
- In the 1950s, notions of the ANN were introduced (Perceptron, McCollough & Pitts)
- In the 1960s, inadequacy of these methods were found (XOR problems), and
- In the 1970s and 1980s, back-propagation provided a reasonable learning approach for multi-layer neural networks for supervised classification problems (Rumelhart & McClelland).
- In the 1970s-1990s, other advances in self-organizing maps, un-supervised learning and hebbian networks provided alternate means for representing knowledge.
- In the 1990s-2000s, other machine learning approaches appeared to take precedence, with lines blurring between ANNs, machine classification, reinforcement learning, and several approaches that linked supervised and unsupervised models (O’Reilly, HTMs, Grossberg).
- In the 2000s, Bayesian approaches were foremost
- In the 2010s, we have seen a resurgence with so-called “deep learnin”g methods. The advances here have been driven by (1) advances in software that allow us to use GPU (graphics cards) to efficiently train and use networks (2) large data labeled data sets, often generated through amazon mechanical turk or as a byproduct of CAPTCHA systems; (3) using 15+ hidden layers; (4) effective use of specific types of layer architectures, including convolutional networks that de-localize patterns from their position in an image.
A simple two-layer neural network can be considered that is essentially what our multinomial regression or logistic regression model is doing. Inputs include a set of features, and output nodes (possibly one per class) are classifications that are learned. Alterately, a pattern can be learned by the output nodes. The main issue here is estimating weights, which are done with heuristic error-propagation approaches rather than MLE or least squares. This is inefficient for two-layer problems, but the heuristic approach will pay off for more complex problems.
The main advance of the late 1970s and early 1980s was adoption of ‘hidden layer’ ANNs. These hidden layers permit solving the XOR problem: when a class is associated with an exclusive or logic. The category structure is stored in a distributed fashion across all nodes, but there is a sense in which the number of hidden nodes controls how complex the classification structure that is possible. With a hidden layer, optimization is difficult with traditional means, but the heuristic approaches generally work reasonably well.
We will start with an image processing example. We will take two images (S and X) and sample features from them to make exemplars:
# library(imager) s <-load.image('s.data.bmp') x <- load.image('x.bmp') svec <-
# as.vector(s) xvec <- as.vector(x) write.csv(svec,'s.csv')
# write.csv(xvec,'x.csv')
## here, the lower the value, the darker the image.
svec <- read.csv("s.csv")
xvec <- read.csv("x.csv")
## reverse the numbers
svec$x <- 255 - svec$x
xvec$x <- 255 - xvec$x
par(mfrow = c(1, 2))
image(matrix(svec$x, 10, 10, byrow = T), col = grey(100:0/100))
image(matrix(xvec$x, 10, 10, byrow = T), col = grey(100:0/100))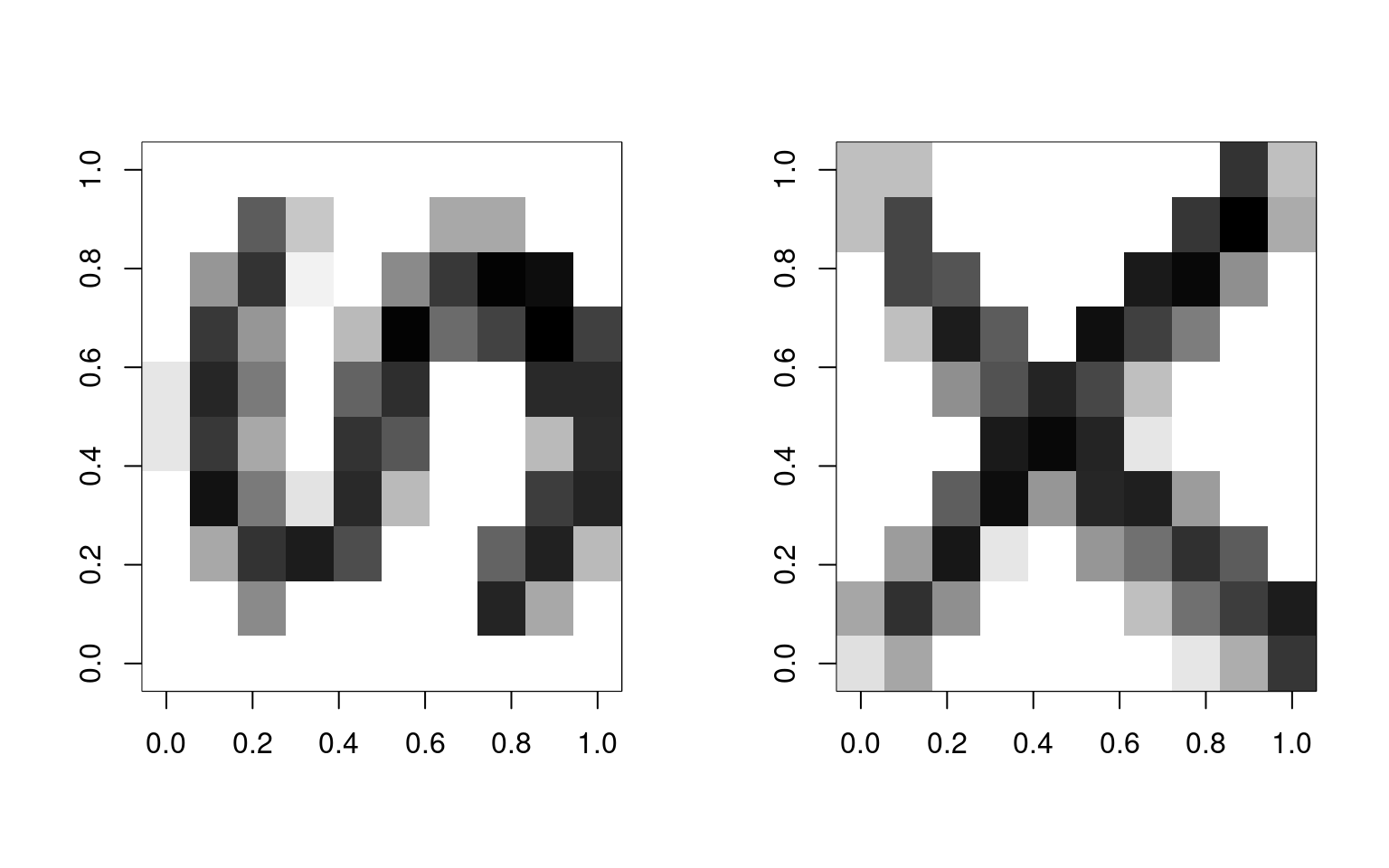 To train the model, we will sample 500 examples of each template:
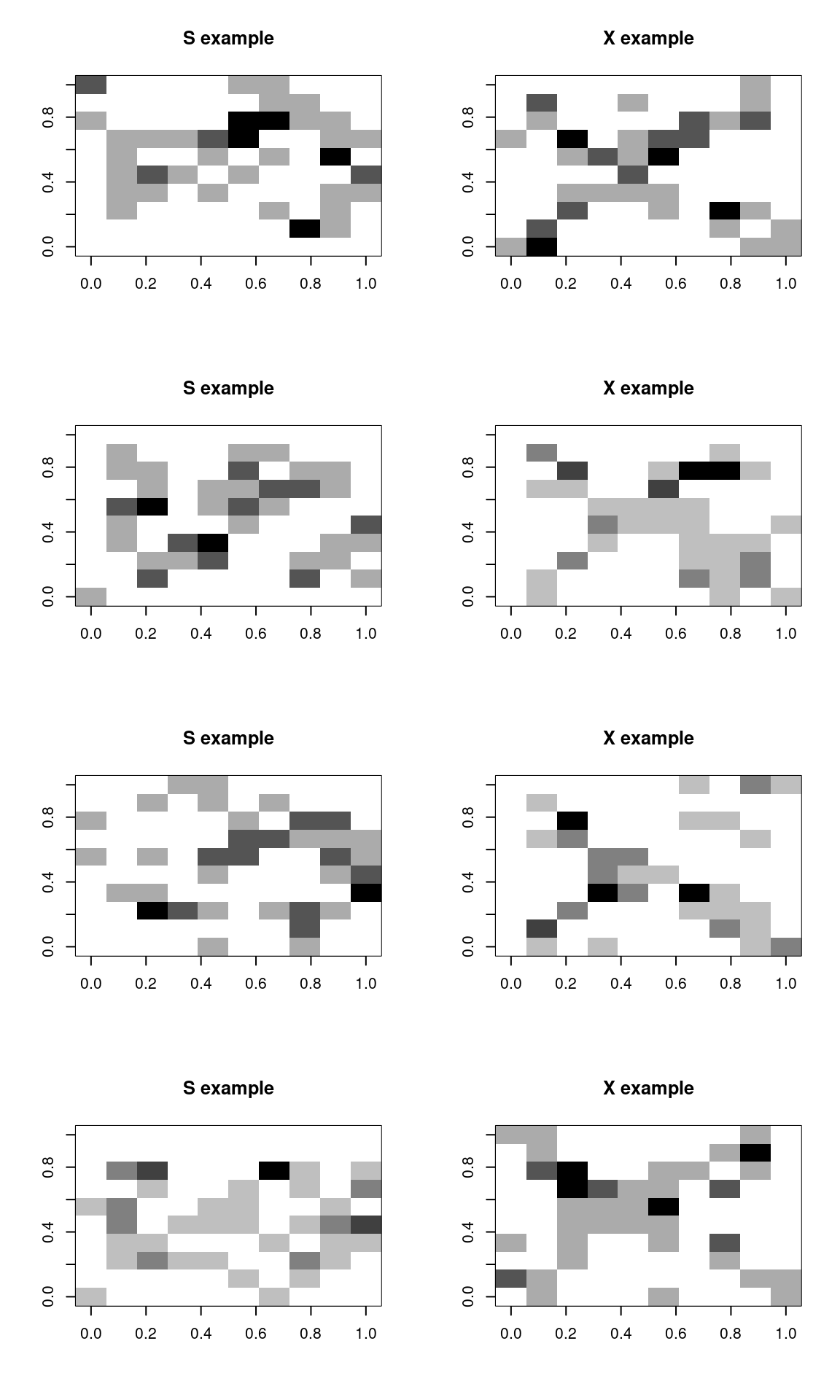
To train the model, we will sample 500 examples of each template:
dataX <- matrix(0, ncol = 100, nrow = 250)
dataS <- matrix(0, ncol = 100, nrow = 250)
letter <- rep(c("x", "s"), each = 250)
par(mfrow = c(4, 2))
for (i in 1:250) {
x <- rep(0, 100)
xtmp <- table(sample(1:100, size = 50, prob = as.matrix(xvec$x), replace = T))
x[as.numeric(names(xtmp))] <- xtmp/max(xtmp)
s <- rep(0, 100)
stmp <- table(sample(1:100, size = 50, prob = as.matrix(svec$x), replace = T))
s[as.numeric(names(stmp))] <- stmp/max(stmp)
if (i <= 4) {
image(matrix(s, 10, 10, byrow = T), main = "S example", col = grey(100:0/100))
image(matrix(x, 10, 10, byrow = T), main = "X example", col = grey(100:0/100))
}
dataX[i, ] <- x
dataS[i, ] <- s
}
Let’s train a neural network on these. Note that we could probably have trained it on the prototypes–maybe from the MNIST library examined in earlier chapters.
library(nnet)
options(warn = 2)
# model <- nnet(letter~data,size=2) #This doesn't work.
# Let's transform the letter to a numeric value
model <- nnet(y = as.numeric(letter == "s"), x = data, size = 2)# weights: 205
initial value 134.702308
final value 0.000000
convergeda 100-2-1 network with 205 weights
options were -# alternately, try using letter as a factor, and use a formula. This also plays
# with some of the learning parameters
merged <- data.frame(letter = as.factor(letter), data)
model2 <- nnet(letter ~ ., data = merged, size = 2, rang = 0.1, decay = 1e-04, maxit = 500,
trace = T)# weights: 205
initial value 346.322926
iter 10 value 0.978601
iter 20 value 0.059554
iter 30 value 0.045672
iter 40 value 0.044185
iter 50 value 0.043833
iter 60 value 0.043422
iter 70 value 0.043135
iter 80 value 0.042890
iter 90 value 0.042723
iter 100 value 0.042618
iter 110 value 0.042548
iter 120 value 0.042533
iter 130 value 0.042527
iter 140 value 0.042525
iter 150 value 0.042525
iter 150 value 0.042525
iter 150 value 0.042525
final value 0.042525
convergedExamining the model
The model contains a lot of information, including some parameters it
is constructed under, and all of the fitted parameters.
We can see when using summary() that we really have two really large
regressions from the 100 input features to 2 hidden nodes, and a single
model with an intercept (b=bias) and two parameters from each hidden
node to the output node. The default is ‘logistic output units’, which
means this last model is essentially a logistic regression.
List of 15
$ n : num [1:3] 100 2 1
$ nunits : int 104
$ nconn : num [1:105] 0 0 0 0 0 0 0 0 0 0 ...
$ conn : num [1:205] 0 1 2 3 4 5 6 7 8 9 ...
$ nsunits : int 104
$ decay : num 0
$ entropy : logi FALSE
$ softmax : logi FALSE
$ censored : logi FALSE
$ value : num 0
$ wts : num [1:205] 6.514 16.642 54.629 -2.507 -0.751 ...
$ convergence : int 0
$ fitted.values: num [1:500, 1] 0 0 0 0 0 0 0 0 0 0 ...
..- attr(*, "dimnames")=List of 2
.. ..$ : NULL
.. ..$ : NULL
$ residuals : num [1:500, 1] 0 0 0 0 0 0 0 0 0 0 ...
..- attr(*, "dimnames")=List of 2
.. ..$ : NULL
.. ..$ : NULL
$ call : language nnet.default(x = data, y = as.numeric(letter == "s"), size = 2)
- attr(*, "class")= chr "nnet"a 100-2-1 network with 205 weights
options were -
b->h1 i1->h1 i2->h1 i3->h1 i4->h1 i5->h1 i6->h1 i7->h1 i8->h1 i9->h1
6.51 16.64 54.63 -2.51 -0.75 -11.67 -19.44 0.27 -4.66 34.21
i10->h1 i11->h1 i12->h1 i13->h1 i14->h1 i15->h1 i16->h1 i17->h1 i18->h1 i19->h1
22.85 42.93 103.99 14.77 -136.22 -106.21 -104.15 -62.49 37.18 96.57
i20->h1 i21->h1 i22->h1 i23->h1 i24->h1 i25->h1 i26->h1 i27->h1 i28->h1 i29->h1
34.94 -1.57 3.97 20.70 24.06 -44.29 -5.45 58.42 -14.75 -95.98
i30->h1 i31->h1 i32->h1 i33->h1 i34->h1 i35->h1 i36->h1 i37->h1 i38->h1 i39->h1
-0.71 -4.40 1.53 -107.80 93.82 114.35 87.75 81.22 -7.13 -32.74
i40->h1 i41->h1 i42->h1 i43->h1 i44->h1 i45->h1 i46->h1 i47->h1 i48->h1 i49->h1
-10.22 -5.16 2.86 -95.27 -59.75 38.84 19.13 -33.20 1.48 1.50
i50->h1 i51->h1 i52->h1 i53->h1 i54->h1 i55->h1 i56->h1 i57->h1 i58->h1 i59->h1
0.89 -3.79 -1.54 59.90 74.24 18.12 -10.47 -5.23 -53.03 -3.33
i60->h1 i61->h1 i62->h1 i63->h1 i64->h1 i65->h1 i66->h1 i67->h1 i68->h1 i69->h1
-0.20 -3.76 30.26 70.65 114.12 9.66 28.29 11.55 9.78 -50.00
i70->h1 i71->h1 i72->h1 i73->h1 i74->h1
-9.48 11.66 -29.55 30.64 50.13
[ reached getOption("max.print") -- omitted 26 entries ]
b->h2 i1->h2 i2->h2 i3->h2 i4->h2 i5->h2 i6->h2 i7->h2 i8->h2 i9->h2
55.36 11.09 39.59 -1.45 0.80 -4.54 -10.06 2.82 -2.64 26.02
i10->h2 i11->h2 i12->h2 i13->h2 i14->h2 i15->h2 i16->h2 i17->h2 i18->h2 i19->h2
19.54 34.08 79.86 14.69 -80.55 -60.74 -58.17 -32.11 35.50 69.24
i20->h2 i21->h2 i22->h2 i23->h2 i24->h2 i25->h2 i26->h2 i27->h2 i28->h2 i29->h2
28.54 0.02 10.45 24.45 24.28 -23.31 6.52 50.81 7.18 -54.88
i30->h2 i31->h2 i32->h2 i33->h2 i34->h2 i35->h2 i36->h2 i37->h2 i38->h2 i39->h2
0.20 -0.41 3.07 -56.86 69.01 83.03 63.17 58.35 -1.15 -17.86
i40->h2 i41->h2 i42->h2 i43->h2 i44->h2 i45->h2 i46->h2 i47->h2 i48->h2 i49->h2
-4.14 -0.41 3.49 -53.29 -28.35 41.55 27.99 -18.03 2.85 1.25
i50->h2 i51->h2 i52->h2 i53->h2 i54->h2 i55->h2 i56->h2 i57->h2 i58->h2 i59->h2
1.96 -0.50 0.02 41.41 57.65 23.97 7.79 14.33 -31.03 0.58
i60->h2 i61->h2 i62->h2 i63->h2 i64->h2 i65->h2 i66->h2 i67->h2 i68->h2 i69->h2
0.83 1.60 23.59 52.50 79.37 9.06 23.07 18.62 20.15 -28.00
i70->h2 i71->h2 i72->h2 i73->h2 i74->h2
-4.23 9.64 -4.37 33.46 37.92
[ reached getOption("max.print") -- omitted 26 entries ]
b->o h1->o h2->o
57.16 -460.68 363.91 Some of the arguments we can control are:
- How the final output classifier works (options involve
linout,entropy,softmax,censored). These control fitting algorithms and approaches, and may impact speed of convergence. The default settings look like they are essentially using least-squared to fit individual nodes. subset: allowing you to train on a subset for cross-validationmask: allowing only some of the input features to be trained.Wts: initial parameter settings. You could train a model on some data, and use those weights to then re-train on new data, for example.decay: this probably controls how far back data in a series are examined. It may allow for a model to adapt to a changing environment better.maxitandtrace: fitting arguments.weights: strength of each case. You may have some cases you want to train on more strongly/often.skip: if TRUE, this is a single-layer network. This is essentially a logistic regression model.
Obtaining predicted response
When we use predict, by default it gives us the ‘raw’ values–activation values returned by the trained network. Because the final layer of this network has just one node (100-2-1), it is just an activation value indicating the class (0 for X and 1 for S). The values are not exactly 0 and 1–they are floating point values very close.
If there were a combined example that was hard to distinguish we would get a different value:
test <- (data[3, ] + data[255, ])/2
par(mfrow = c(1, 3))
image(matrix(data[3, ], 10), col = grey(100:0/100))
image(matrix(data[255, ], 10), col = grey(100:0/100))
image(matrix(test, 10), col = grey(100:0/100))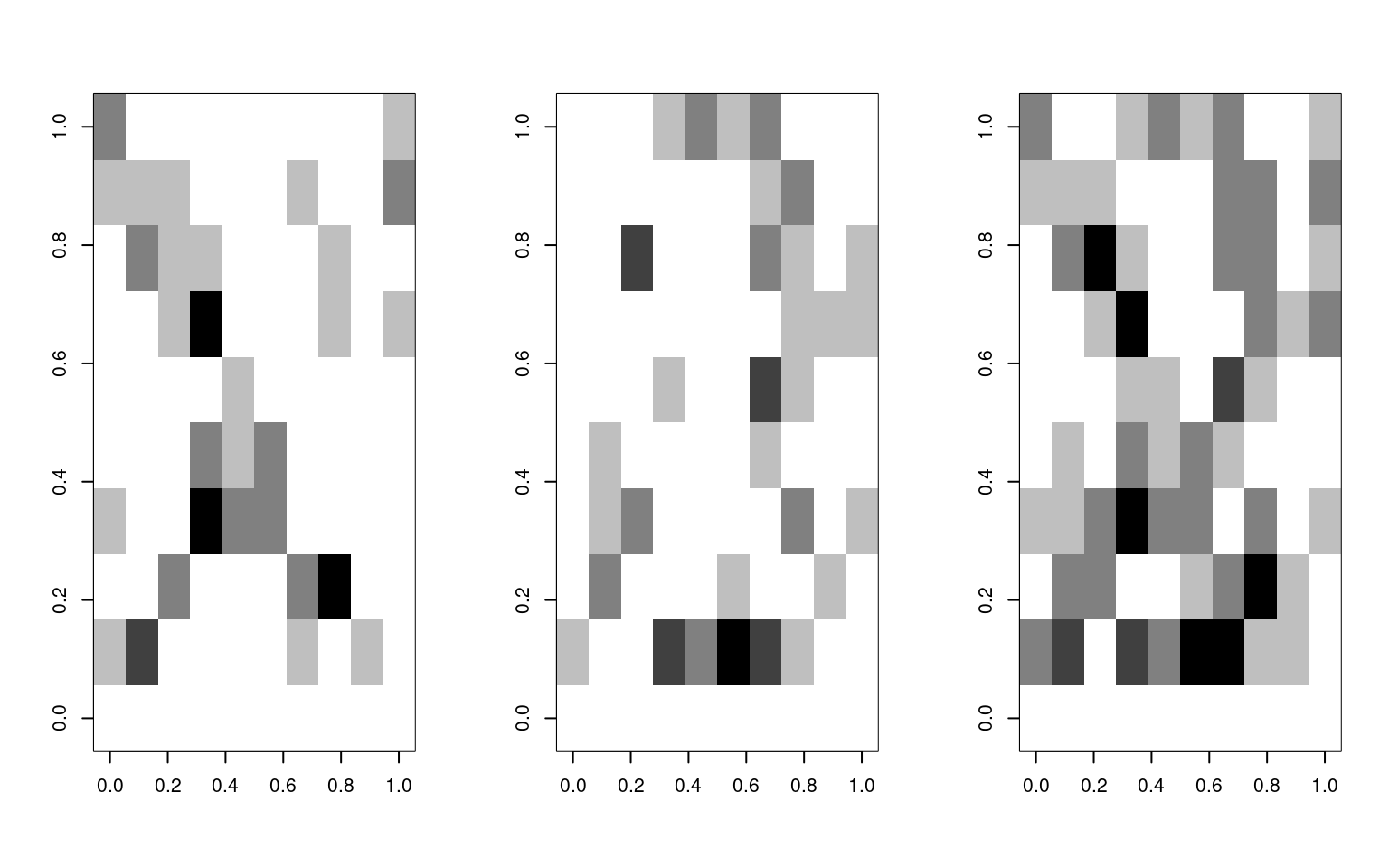
[,1]
[1,] 0 [,1]
[1,] 1 [,1]
[1,] 1In this case, the X shows up as a strong X, the S shows up as an S, but the combined version is a slightly weaker S.
We can get classifications using type=“class”:
[1] "x" "x" "x" "x" "x" "x" "x" "x" "x" "x" "x" "x" "x" "x" "x" "x" "x" "x" "x"
[20] "x" "x" "x" "x" "x" "x" "x" "x" "x" "x" "x" "x" "x" "x" "x" "x" "x" "x" "x"
[39] "x" "x" "x" "x" "x" "x" "x" "x" "x" "x" "x" "x" "x" "x" "x" "x" "x" "x" "x"
[58] "x" "x" "x" "x" "x" "x" "x" "x" "x" "x" "x" "x" "x" "x" "x" "x" "x" "x"
[ reached getOption("max.print") -- omitted 425 entries ]
letter s x
s 250 0
x 0 250Training with very limited/noisy examples
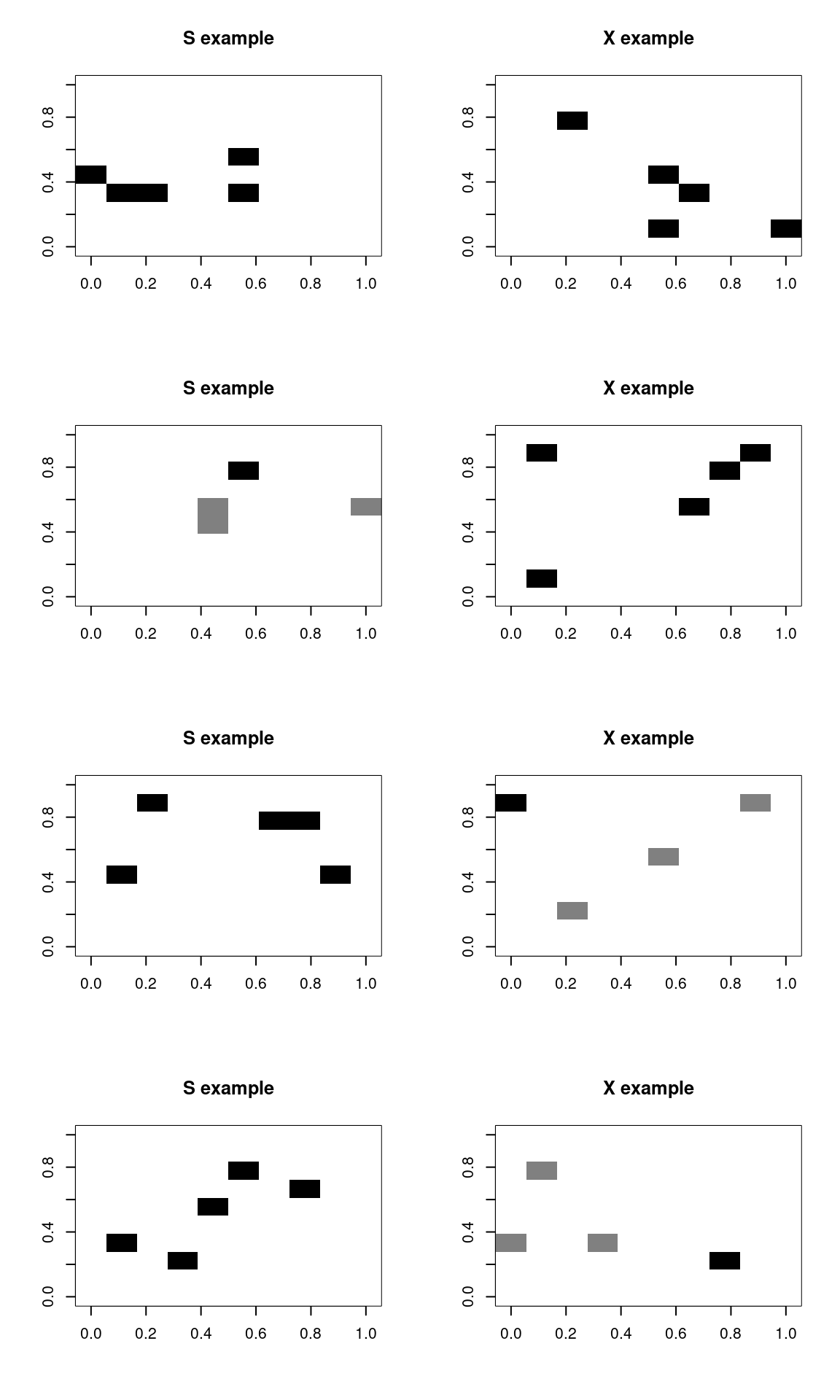
This classification is actually perfect, but there was a lot of information available. Let’s sample just 5 points out to create the pattern:
dataX <- matrix(0, ncol = 100, nrow = 250)
dataS <- matrix(0, ncol = 100, nrow = 250)
letter <- rep(c("x", "s"), each = 250)
par(mfrow = c(4, 2))
for (i in 1:250) {
x <- rep(0, 100)
xtmp <- table(sample(1:100, size = 5, prob = as.matrix(xvec$x), replace = T))
x[as.numeric(names(xtmp))] <- xtmp/max(xtmp)
s <- rep(0, 100)
stmp <- table(sample(1:100, size = 5, prob = as.matrix(svec$x), replace = T))
s[as.numeric(names(stmp))] <- stmp/max(stmp)
## plot the first few examples:
if (i <= 4) {
image(matrix(s, 10, 10, byrow = T), main = "S example", col = grey(100:0/100))
image(matrix(x, 10, 10, byrow = T), main = "X example", col = grey(100:0/100))
}
dataX[i, ] <- x
dataS[i, ] <- s
}
data <- rbind(dataX, dataS)
merged <- data.frame(letter = as.factor(letter), data)
model3 <- nnet(letter ~ ., data = merged, size = 2, rarg = 0.1, decay = 1e-04, maxit = 500)# weights: 205
initial value 349.342458
iter 10 value 98.547003
iter 20 value 45.166893
iter 30 value 30.523416
iter 40 value 28.985007
iter 50 value 28.355796
iter 60 value 27.688482
iter 70 value 26.956694
iter 80 value 26.550998
iter 90 value 24.582976
iter 100 value 24.418685
iter 110 value 24.330491
iter 120 value 24.206411
iter 130 value 23.996977
iter 140 value 23.323214
iter 150 value 19.973030
iter 160 value 16.959922
iter 170 value 14.900277
iter 180 value 14.703310
iter 190 value 14.631866
iter 200 value 14.591938
iter 210 value 14.543914
iter 220 value 14.432881
iter 230 value 14.355548
iter 240 value 14.329130
iter 250 value 14.301357
iter 260 value 14.187941
iter 270 value 14.062408
iter 280 value 14.031454
iter 290 value 14.021345
iter 300 value 14.014148
iter 310 value 14.007568
iter 320 value 14.000887
iter 330 value 13.931244
iter 340 value 13.867128
iter 350 value 13.827783
iter 360 value 7.780889
iter 370 value 7.504392
iter 380 value 7.436209
iter 390 value 7.396963
iter 400 value 7.350464
iter 410 value 7.309284
iter 420 value 7.276907
iter 430 value 7.246058
iter 440 value 7.222203
iter 450 value 7.201361
iter 460 value 7.191541
iter 470 value 7.185425
iter 480 value 7.178740
iter 490 value 7.172972
iter 500 value 7.162451
final value 7.162451
stopped after 500 iterations
letter s x
s 250 0
x 1 249It still does very well, with a few errors, for very sparse data. In fact, it might be doing TOO well. That is, in a sense, it is picking up on arbitrary but highly diagnostic features. A human observer would never be confident in the outcome, because the information is too sparse, but in this limited world, a single pixel is enough to make a good guess.
Neural Networks and the XOR problem
Neural networks became popular when researchers realized that networks with a hidden layer could solve ‘XOR classification problems’. Early on, researchers recognized that a simple perception (2-layer) neural network could be used for AND or OR combinations, but not XOR, as these are not linearly separable. XOR classification maps onto many real-world interaction problems. For example, two safe pharmaceuticals might be dangerous when taken together, and a simple neural network could never detect this state–if one is good, and the other is good, both must be better. An XOR problem is one in which one feature or another (but not both or neither) indicate class membership. In order to perform classification with this logic, a hidden layer is required.
Here is a class defined by an XOR structure:
library(MASS)
library(DAAG)
feature1 <- rnorm(200)
feature2 <- rnorm(200)
outcome <- as.factor((feature1 > 0.6 & feature2 > 0.3) | (feature1 > 0.6 & feature2 <
0.3))
outcome <- as.factor((feature1 * (-feature2) + rnorm(200)) > 0)The linear discriminant model fails to discriminate (at least without an interaction)
Overall accuracy = 0.6
Confusion matrix
Predicted (cv)
Actual FALSE TRUE
FALSE 0.387 0.613
TRUE 0.215 0.785Overall accuracy = 0.695
Confusion matrix
Predicted (cv)
Actual FALSE TRUE
FALSE 0.602 0.398
TRUE 0.224 0.776Similarly, the neural networks with an empty single layer (skip=TRUE) are not great at discriminating, but with a few hidden nodes they work well. Note that the size argument essentially overrides the skip argument, as n2 and n3 are both 2-3-1 networks:
# weights: 3
initial value 153.137485
final value 136.081429
convergedOverall accuracy = 0.4
Confusion matrix
Predicted (cv)
Actual FALSE TRUE
FALSE 0.613 0.387
TRUE 0.785 0.215# weights: 15
initial value 149.958156
iter 10 value 107.371535
iter 20 value 105.720295
iter 30 value 104.412357
iter 40 value 104.161983
iter 50 value 104.043551
iter 60 value 103.894953
iter 70 value 103.831541
iter 80 value 103.562693
iter 90 value 103.424503
iter 100 value 103.399276
final value 103.399276
stopped after 100 iterationsOverall accuracy = 0.695
Confusion matrix
Predicted (cv)
Actual FALSE TRUE
FALSE 0.774 0.226
TRUE 0.374 0.626# weights: 13
initial value 139.045510
iter 10 value 126.944698
iter 20 value 113.676168
iter 30 value 110.715150
iter 40 value 109.930469
iter 50 value 108.405803
iter 60 value 107.109966
iter 70 value 106.496786
iter 80 value 105.787198
iter 90 value 105.646894
iter 100 value 105.590004
final value 105.590004
stopped after 100 iterationsOverall accuracy = 0.68
Confusion matrix
Predicted (cv)
Actual FALSE TRUE
FALSE 0.720 0.280
TRUE 0.355 0.645Because we have the XOR structure, the simple neural network without a hidden layer is essentially LDA, and in fact often gets a similar level of accuracy (53%).
IPhone Data using the NNet
To model the iphone data set, we need to decide on how large of a model we want. We need to also remember that training a model like this is not deterministic–every time we do it the situation will be a little different. Because we have just two classes, maybe a 2-layer hidden network would work.
By fitting the model several times, we can see that it performs differently every time. At times, the model does reasonably well. This does about as well as any of the best classification models. Curiously, this particular model has a bias toward Android accuracy.
phone <- read.csv("data_study1.csv")
phone$Smartphone <- (as.factor(phone$Smartphone))
phone$Gender <- as.numeric(as.factor(phone$Gender))
set.seed(100)
phonemodel <- nnet(Smartphone ~ ., size = 3, data = phone)# weights: 43
initial value 368.669183
iter 10 value 351.629336
iter 20 value 328.109320
iter 30 value 319.345472
iter 40 value 317.075499
iter 50 value 314.039955
iter 60 value 313.953479
final value 313.953382
convergedconfusion(phone$Smartphone, factor(predict(phonemodel, newdata = phone, type = "class",
levels = c("Android", "iPhone"))))Overall accuracy = 0.652
Confusion matrix
Predicted (cv)
Actual Android iPhone
Android 0.603 0.397
iPhone 0.313 0.687Other times, it does poorly: Here, it calls everything an iPhone:
# weights: 29
initial value 457.289985
final value 358.808683
convergedconfusion(phone$Smartphone, factor(predict(phonemodel2, newdata = phone, type = "class"),
levels = c("Android", "iPhone")))Overall accuracy = 0.586
Confusion matrix
Predicted (cv)
Actual Android iPhone
Android 0 1
iPhone 0 1In this case, it called everything in iPhone, resulting in 58.6% accuracy. Like many of our approaches, we are doing heuristic optimization and so may end up in local optima. It is thus useful to run the model many several times and look for the best model. Running this many times, the best models seem to get around 370-390 correct, which is in the low 70% for accuracy.
preds <- matrix(0, 100)
for (i in 1:100) {
phonemodel3 <- nnet(Smartphone ~ ., data = phone, size = 2)
preds[i] <- confusion(phone$Smartphone, factor(predict(phonemodel3, newdata = phone,
type = "class"), levels = c("Android", "iPhone")))$overall
}
hist(preds, col = "gold", main = "Accuracy of 100 fitted models (2 hidden nodes)",
xlim = c(0, 1))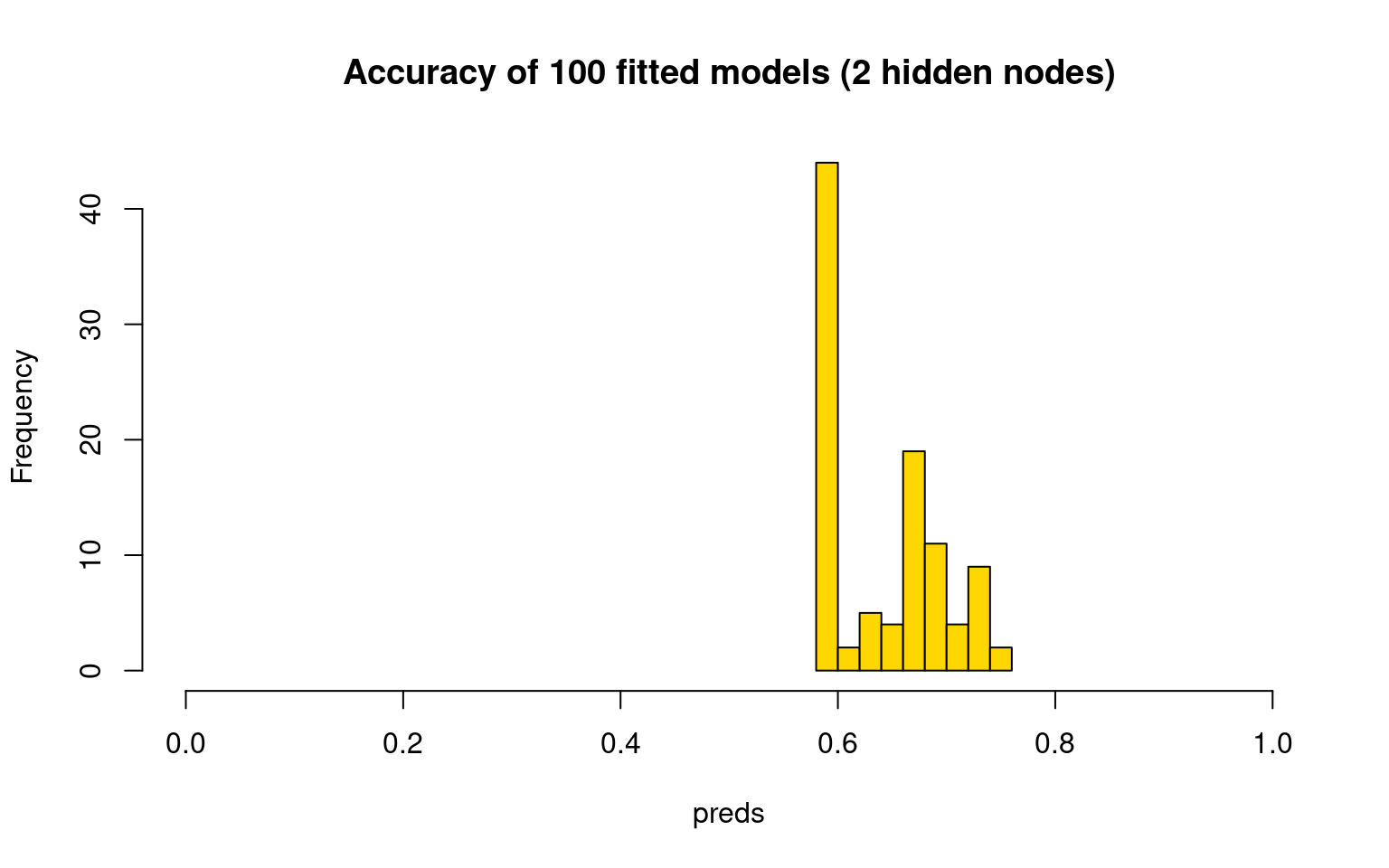 Do we do any better, on average, with more hidden nodes?
Do we do any better, on average, with more hidden nodes?
preds <- matrix(0, 100)
for (i in 1:100) {
phonemodel3 <- nnet(Smartphone ~ ., data = phone, size = 6, trace = FALSE)
# preds[i] <-
# confusion(phone$Smartphone,factor(predict(phonemodel3,newdata=phone,type='class'),levels=c('Android','iPhone')))$overall
tab <- table(phone$Smartphone, factor(predict(phonemodel3, newdata = phone, type = "class"),
levels = c("Android", "iPhone")))
preds[i] <- sum(diag(tab))/sum(tab)
}
hist(preds, col = "gold", main = "Accuracy of 100 fitted models (6 hidden nodes)",
xlim = c(0, 1))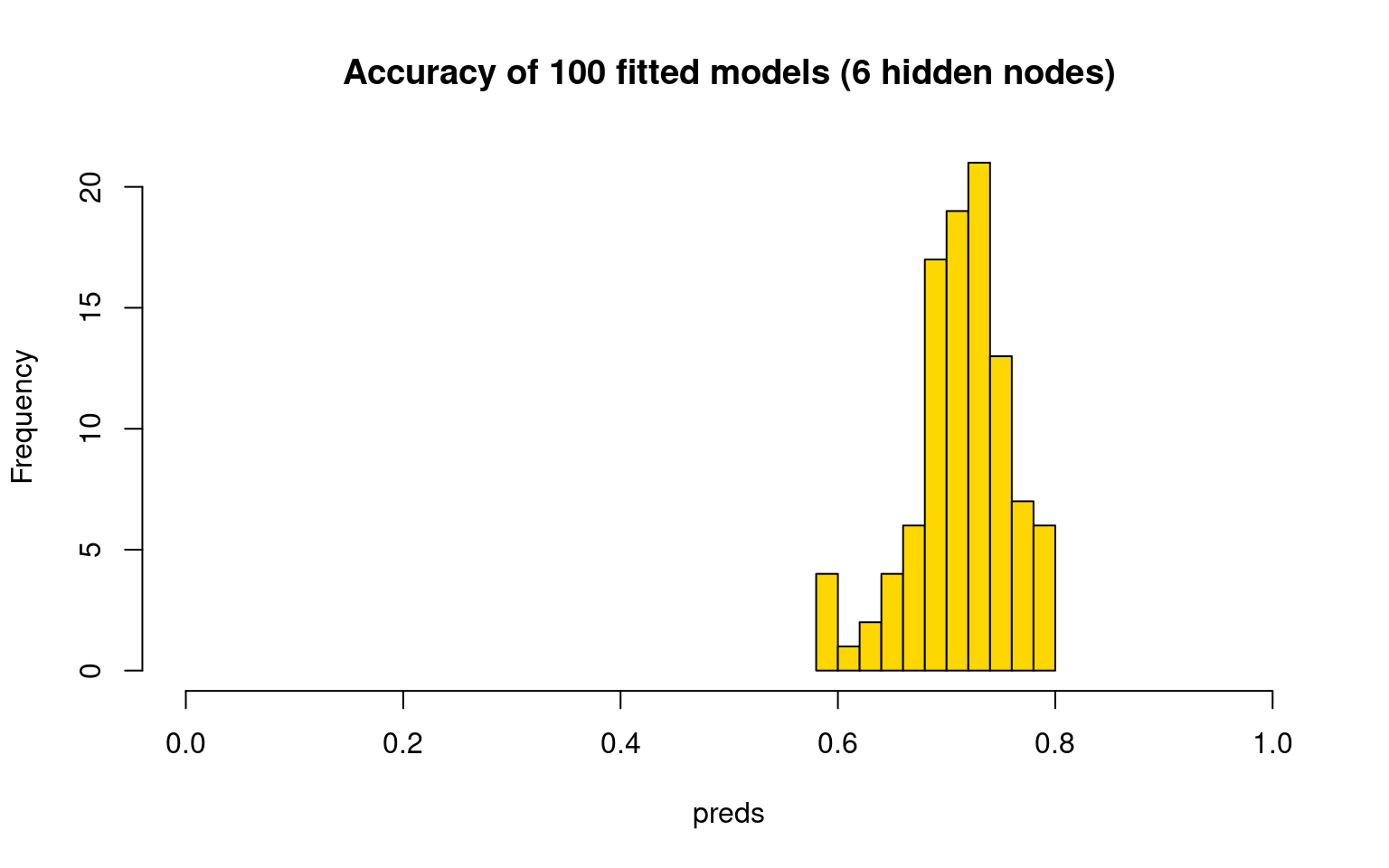 This seems to be more consistent, and do better overall–usually above
70% accuracy. But like every model we have examined, the best models are
likely to be over-fitting, and getting lucky at re-predicting their own
data. It would also be important to implement a cross-validation scheme.
On its own, it might just tell us if we are overfitting. We could use it
to help select the number of hidden nodes, or to select variables for
exclusion.
This seems to be more consistent, and do better overall–usually above
70% accuracy. But like every model we have examined, the best models are
likely to be over-fitting, and getting lucky at re-predicting their own
data. It would also be important to implement a cross-validation scheme.
On its own, it might just tell us if we are overfitting. We could use it
to help select the number of hidden nodes, or to select variables for
exclusion.
There is no built-in cross-validation here, but you can implement one using subset functions.
train <- rep(FALSE, nrow(phone))
train[sample(1:nrow(phone), size = 300)] <- TRUE
test <- !train
phonemodel2 <- nnet(Smartphone ~ ., data = phone, size = 6, subset = train)# weights: 85
initial value 204.327276
iter 10 value 193.352724
iter 20 value 166.957659
iter 30 value 136.845896
iter 40 value 126.652225
iter 50 value 118.844038
iter 60 value 114.601579
iter 70 value 112.362718
iter 80 value 109.187074
iter 90 value 106.922061
iter 100 value 104.486444
final value 104.486444
stopped after 100 iterationsOverall accuracy = 0.563
Confusion matrix
Predicted (cv)
Actual Android iPhone
Android 0.340 0.660
iPhone 0.273 0.727We can try this 100 times and see how well it does on the cross-validation set:
preds <- rep(0, 100)
for (i in 1:100) {
train <- rep(FALSE, nrow(phone))
train[sample(1:nrow(phone), size = 300)] <- TRUE
test <- !train
phonemodel2 <- nnet(Smartphone ~ ., data = phone, size = 6, subset = train, trace = FALSE)
tab <- table(phone$Smartphone[test], factor(predict(phonemodel2, newdata = phone[test,
], type = "class"), levels = c("Android", "iPhone")))
preds[i] <- sum(diag(tab))/sum(tab)
# preds[i] <-
# confusion(phone$Smartphone[test],factor(predict(phonemodel2,newdata=phone[test,],type='class'),levels=c('Android','iPhone')))$overall
}
hist(preds, col = "gold", main = "Accuracy of 100 cross-validated models (6 hidden nodes)",
xlim = c(0, 1))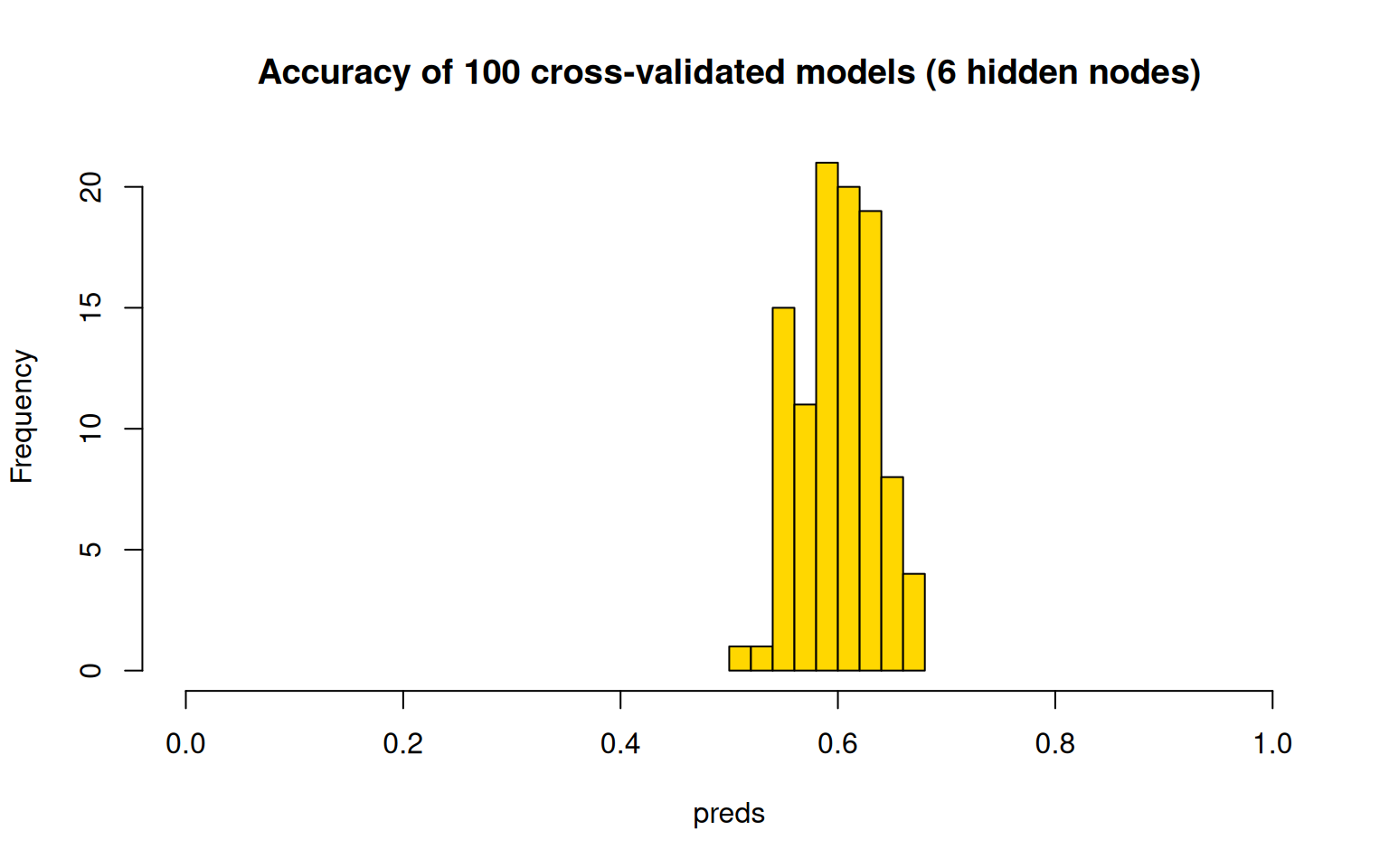
The cross-validation scores are typically a bit lower, with models getting around 60% on average, and up to 70% for the best. Now that we have built this, we could use average cross-validation accuracy to help select variables for exclusion. Here, let’s just test the predictors we have found previously to be fairly good:
preds <- rep(0, 100)
for (i in 1:100) {
train <- rep(FALSE, nrow(phone))
train[sample(1:nrow(phone), size = 300)] <- TRUE
test <- !train
phonemodel2 <- nnet(Smartphone ~ Gender + Avoidance.Similarity + Phone.as.status.object +
Age, data = phone, size = 6, subset = train, trace = F)
tab <- table(phone$Smartphone[test], factor(predict(phonemodel2, newdata = phone[test,
], type = "class"), levels = c("Android", "iPhone")))
preds[i] <- sum(diag(tab))/sum(tab)
}
hist(preds, col = "gold", main = "Accuracy of 100 cross-validated models (6 hidden nodes)",
xlim = c(0, 1))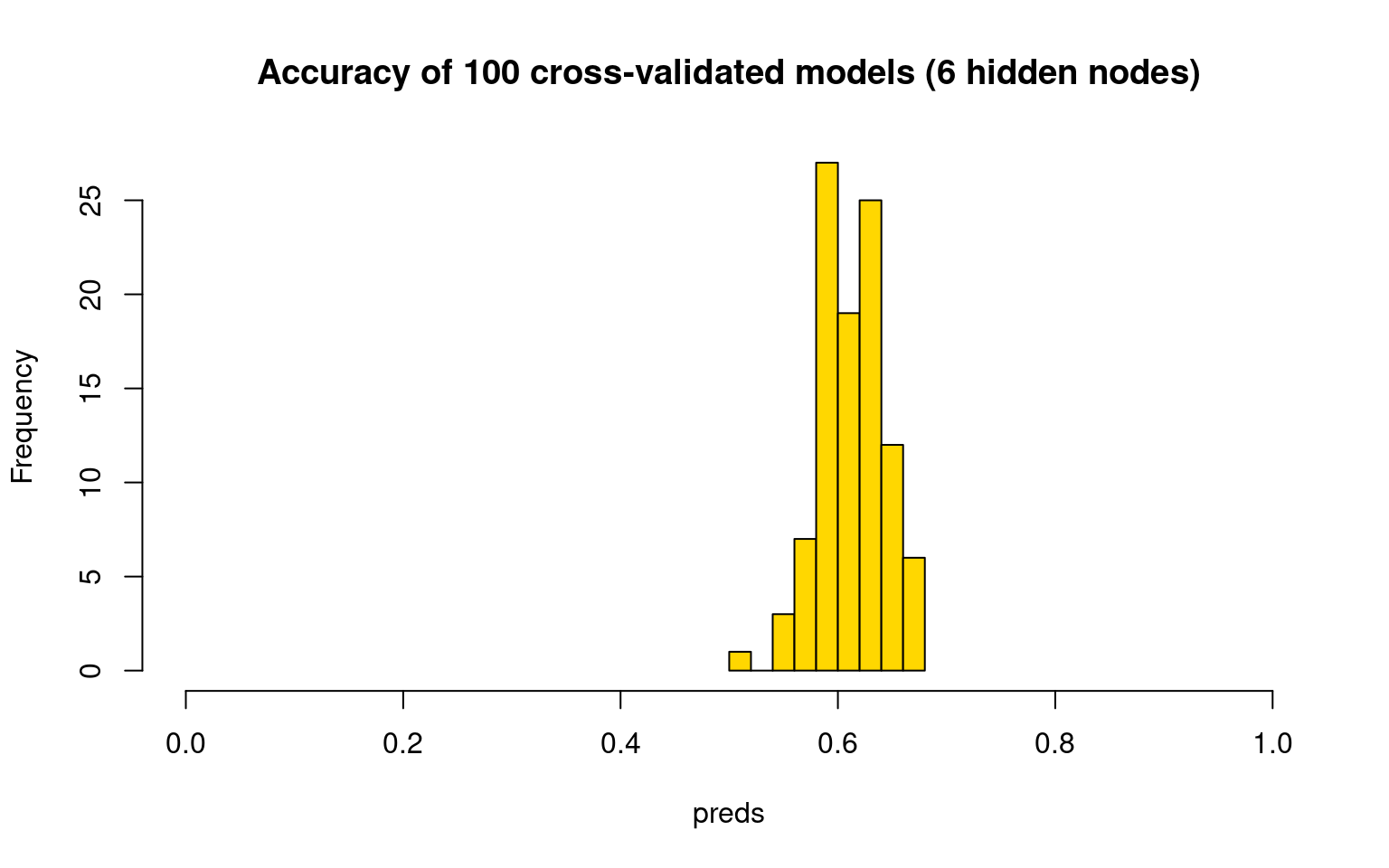
Most of these are better than chance, and it seems to do about as well as the full set of predictors as well.
The neuralnet library
The neuralnet library is perhaps a more flexible implementation, with multiple hidden layers. Here we have 2 hidden layers with two nodes each. When fitting it, it seemed to get stuck frequently and not converge or have some other error, but it does give a nice graphical visualization of the network. It seems to used more advanced backprop algorithms and activation functions. Here we have two hidden layers with two nodes each. The network does not make a lot of sense, but the model fails to converge under many larger network conditions.
library(neuralnet)
set.seed(10313)
train <- rep(FALSE, nrow(phone))
train[sample(1:nrow(phone), size = 300)] <- TRUE
test <- !train
phonemodel3 <- neuralnet(Smartphone ~ ., hidden = c(2, 2), data = phone[train, ])
pred <- apply(predict(phonemodel3, newdata = phone[test, ]), 1, which.max)
ptest <- phone[test, ]
acc <- (sum(ptest[pred == 1, ]$Smartphone == "Android") + sum(ptest[pred == 2, ]$Smartphone ==
"iPhone"))/sum(test)
print(acc)[1] 0.5764192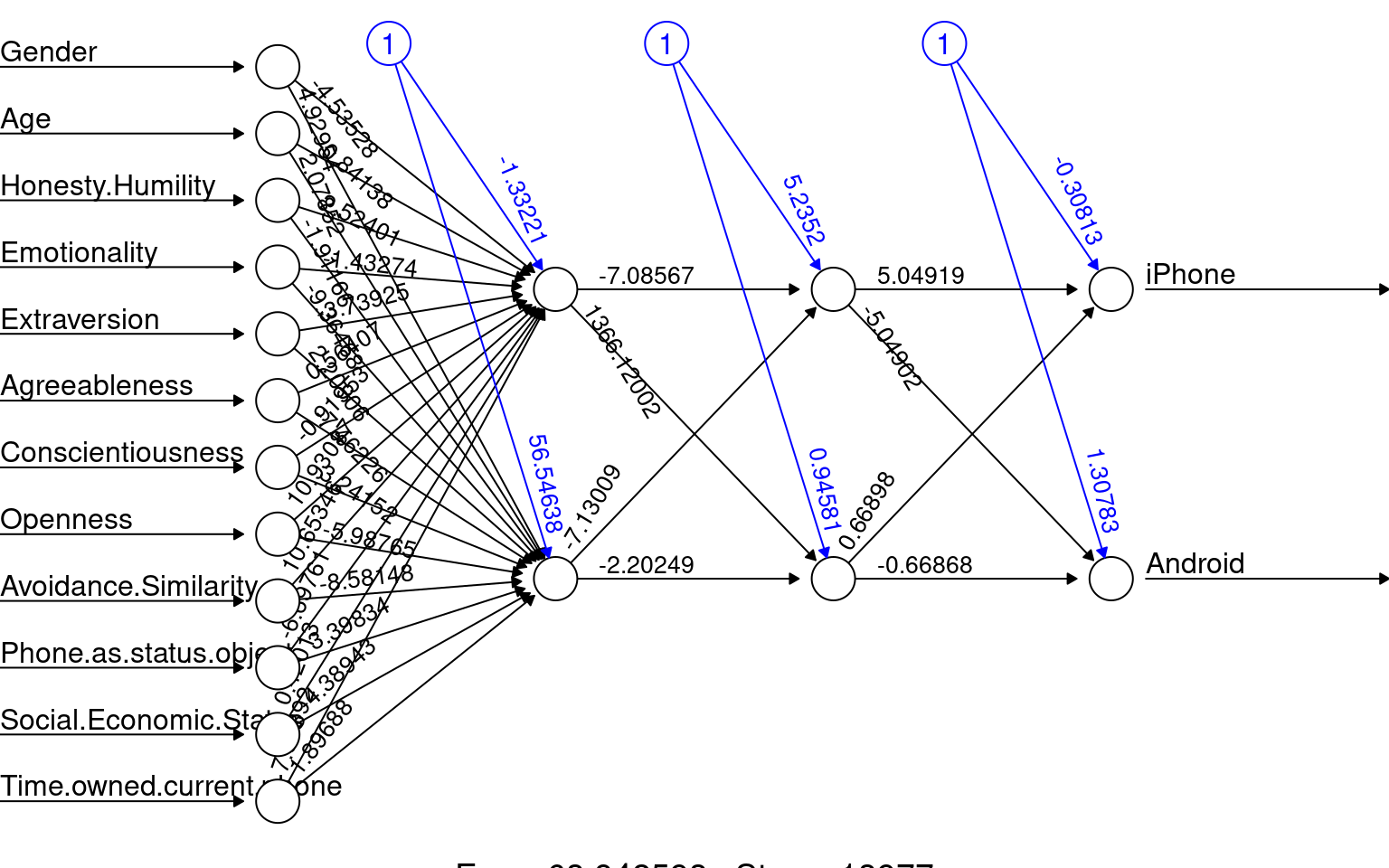
Classifying handwritten characters using a neural net
The neuralnet library is a bit more powerful, and allows
multiple layers. Let’s try it for a larger data set–mnist handwritten
characters. I’ve included 10,000 cases inn mnist_test.csv, but this gets
a bit difficult to build a model, so let’s consider three characters
that are similar: 4, 7, and 9. This results in about 3000 cases, so lets
fit the data on a subset of 1000, using 10 hidden nodes.
library(neuralnet)
library(DAAG)
## 10K examples of 0 to 9
mnistdat <- read.csv("mnist_test.csv")
mnistdat$label <- factor(mnistdat$label) ## this needs to be a factor if it has more than 2 levels
mnist47 <- mnistdat[mnistdat$label == 4 | mnistdat$label == 7 | mnistdat$label ==
9, ]
train <- mnist47[sample(1:nrow(mnist47), 1000), ] #train on some examples
set.seed(100)
mnist <- neuralnet(label ~ ., hidden = c(4), data = train)
## examine predictions on the the trained data:
table(train$label, c(4, 7, 9)[apply(mnist$response, 1, which.max)])
4 7 9
0 0 0 0
1 0 0 0
2 0 0 0
3 0 0 0
4 319 0 0
5 0 0 0
6 0 0 0
7 0 358 0
8 0 0 0
9 0 0 323It essentially predicts the trained data perfectly. Let’s see how it
does on the held-out data. We will use the predict function
to get the activation values, and then use the which.max
function to get the predicted class. We will then compare the predicted
class to the actual class.
## examine held-out data:
p2 <- predict(mnist, newdata = mnist47)
pred <- c(4, 7, 9)[apply(p2, 1, which.max)]
table(mnist47$label, pred) pred
4 7 9
0 0 0 0
1 0 0 0
2 0 0 0
3 0 0 0
4 654 10 318
5 0 0 0
6 0 0 0
7 17 906 105
8 0 0 0
9 72 36 901[1] 0.8151706Errors of 4 categorized as a 7:
error47 <- mnist47[mnist47$label == 4 & pred == 7, ]
cols <- grey(100:1/100)
par(mfrow = c(2, 4))
image((matrix(as.numeric(error47[1, -1]), nrow = 28, ncol = 28))[, 28:1], xaxt = "n",
yaxt = "n", col = cols, main = "4s called 7s")
image((matrix(as.numeric(error47[2, -1]), nrow = 28, ncol = 28))[, 28:1], xaxt = "n",
yaxt = "n", col = cols, main = "4s called 7s")
image((matrix(as.numeric(error47[3, -1]), nrow = 28, ncol = 28))[, 28:1], xaxt = "n",
yaxt = "n", col = cols, main = "4s called 7s")
image((matrix(as.numeric(error47[4, -1]), nrow = 28, ncol = 28))[, 28:1], xaxt = "n",
yaxt = "n", col = cols, main = "4s called 7s")
image((matrix(as.numeric(error47[5, -1]), nrow = 28, ncol = 28))[, 28:1], xaxt = "n",
yaxt = "n", col = cols, main = "4s called 7s")
image((matrix(as.numeric(error47[6, -1]), nrow = 28, ncol = 28))[, 28:1], xaxt = "n",
yaxt = "n", col = cols, main = "4s called 7s")
image((matrix(as.numeric(error47[7, -1]), nrow = 28, ncol = 28))[, 28:1], xaxt = "n",
yaxt = "n", col = cols, main = "4s called 7s")
image((matrix(as.numeric(error47[8, -1]), nrow = 28, ncol = 28))[, 28:1], xaxt = "n",
yaxt = "n", col = cols, main = "4s called 7s")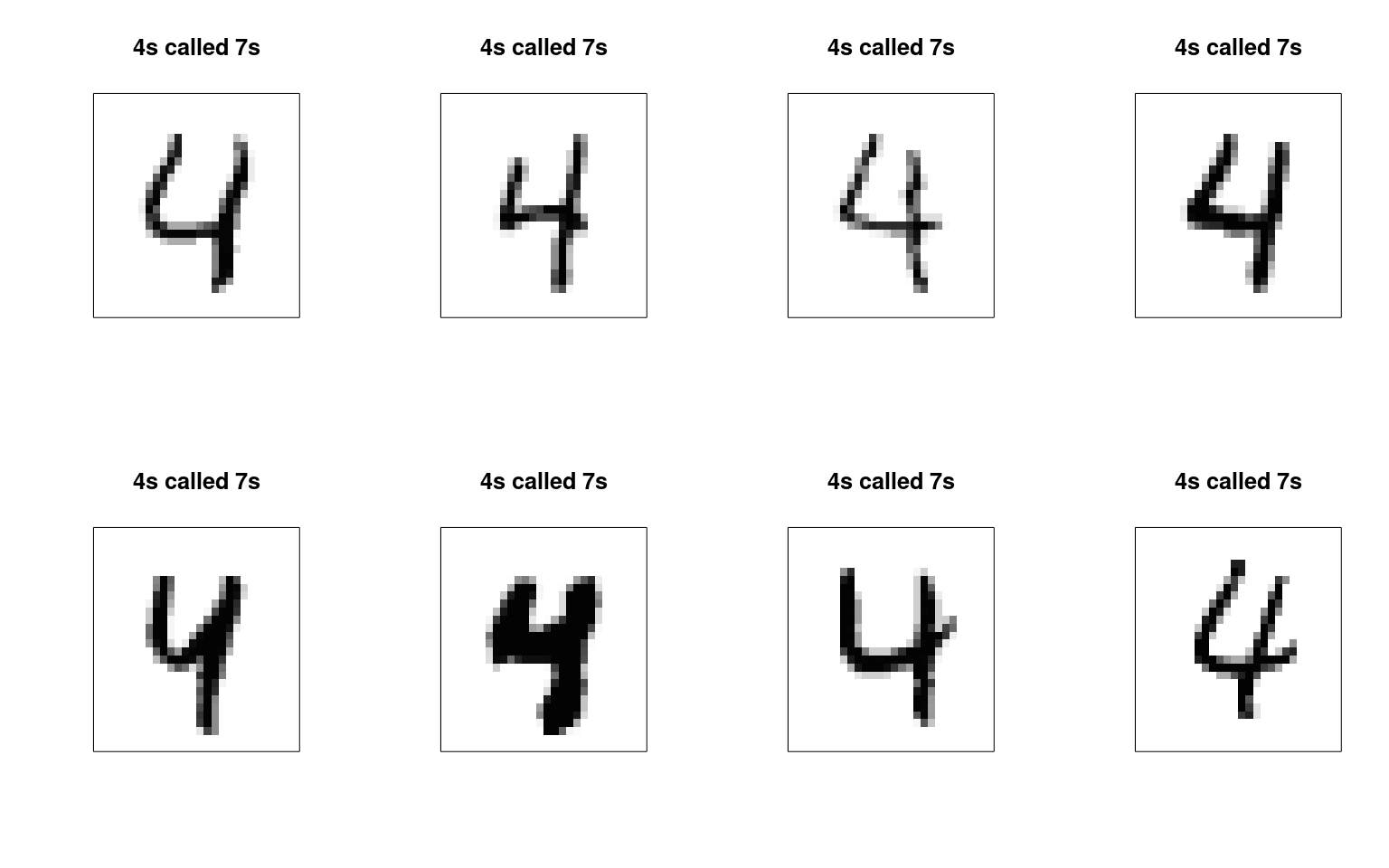
Errors of 4 categorized as a 9:
error49 <- mnist47[mnist47$label == 4 & pred == 9, ]
cols <- grey(100:1/100)
par(mfrow = c(2, 4))
image((matrix(as.numeric(error49[1, -1]), nrow = 28, ncol = 28))[, 28:1], xaxt = "n",
yaxt = "n", col = cols, main = "4s called 7s")
image((matrix(as.numeric(error49[2, -1]), nrow = 28, ncol = 28))[, 28:1], xaxt = "n",
yaxt = "n", col = cols, main = "4s called 7s")
image((matrix(as.numeric(error49[3, -1]), nrow = 28, ncol = 28))[, 28:1], xaxt = "n",
yaxt = "n", col = cols, main = "4s called 7s")
image((matrix(as.numeric(error49[4, -1]), nrow = 28, ncol = 28))[, 28:1], xaxt = "n",
yaxt = "n", col = cols, main = "4s called 7s")
image((matrix(as.numeric(error49[5, -1]), nrow = 28, ncol = 28))[, 28:1], xaxt = "n",
yaxt = "n", col = cols, main = "4s called 7s")
image((matrix(as.numeric(error49[6, -1]), nrow = 28, ncol = 28))[, 28:1], xaxt = "n",
yaxt = "n", col = cols, main = "4s called 7s")
image((matrix(as.numeric(error49[7, -1]), nrow = 28, ncol = 28))[, 28:1], xaxt = "n",
yaxt = "n", col = cols, main = "4s called 7s")
image((matrix(as.numeric(error49[8, -1]), nrow = 28, ncol = 28))[, 28:1], xaxt = "n",
yaxt = "n", col = cols, main = "4s called 7s") We can maybe see some patterns here for what kinds of errors are being
made.
We can maybe see some patterns here for what kinds of errors are being
made.
This is just the tip of the iceberg for neural network models. Modern deep networks use a mixture of different kinds of sublayers that are specialized for handling specific kinds of data (e.g., recurrent, convolutional, etc.), and they also use 15+ layers. These require substantial compute power to fit, and huge data sets of labeled data–often labeled by humans. But this is the basis for many of the recent advances in artificial intelligence.